
Chemoinformatics tools
Institute of Molecular and Translational Medicine, Palacky University
CReM: chemically reasonable mutations framework
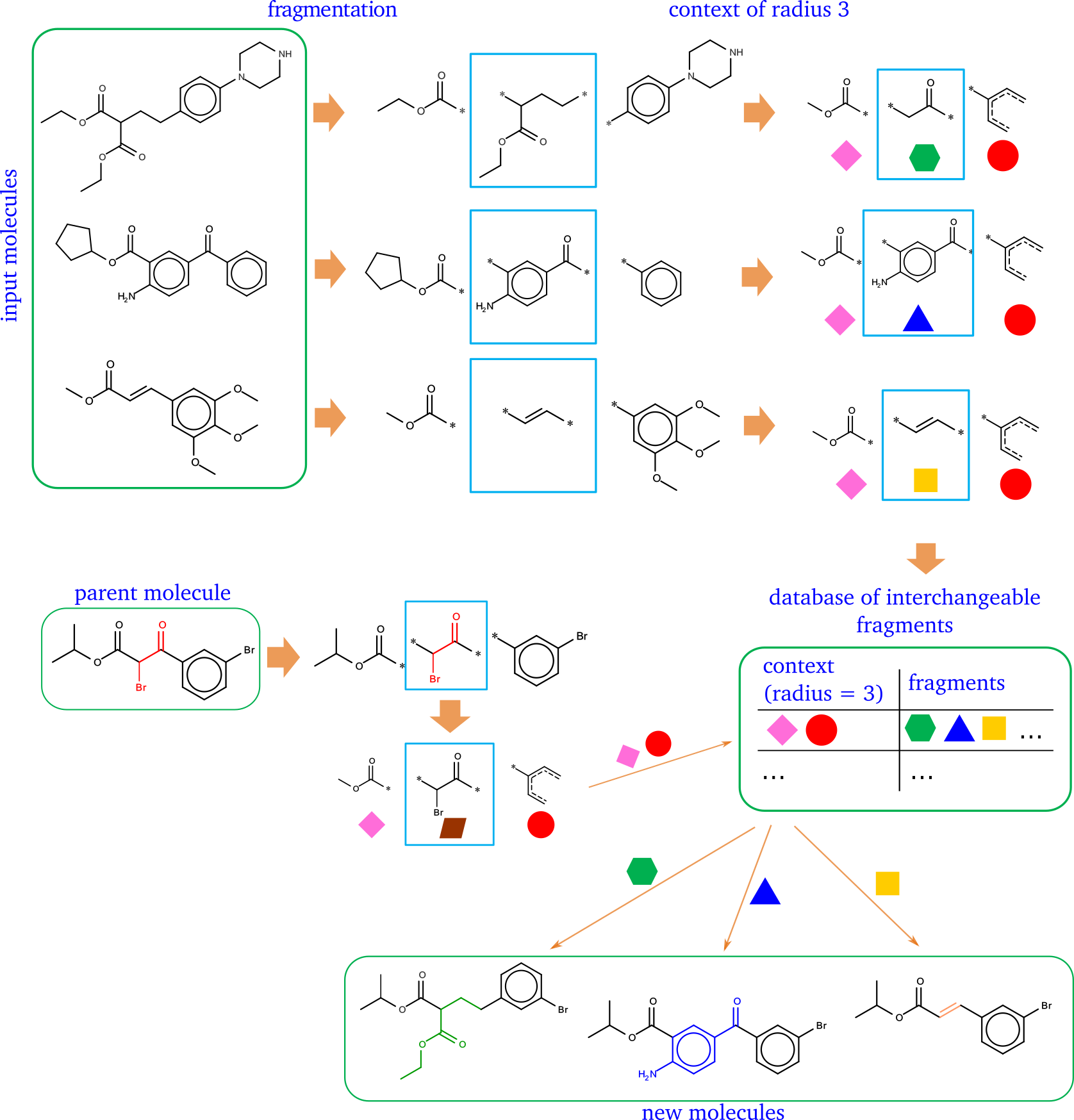
The open-source framework for fragment-based generation of chemical structures. The idea is similar to matched molecular pair - if two fragments are in the identical contexts they can replace each other to produce new chemically valid and probably synthetically feasible structures.
Features:
- Generation of chemically valid structures
- Control of chemotypes of generated structure by choosing a radius of a chemical context considered during replacement. No new fragments which are absent in a fragment database and have the size equal or less then the specified context radius will appear in generated structures.
- Indirect control of synthetic accessibility of generated structure by managment of the content of a fragment database. A database created from more synthetically accessibe compounds will result in more synthetically accessibe structures.
Implementation features:
- Generation of a custom fragment database
- Three modes of structure generation: MUTATE, GROW, LINK
- Context radius to consider for replacement
- Selection of the fragment size to replace and the size of a replacing fragment
- Protection of atoms from modification (e.g. scaffold protection)
- Replacements with fragments occurred in a fragment database with certain minimal frequency
- Make randomly chosen replacements up to the specified number
Repository:
https://github.com/DrrDom/cremLinks to fragment databases:
These databases were created using CReM 0.2 and RDKit 2019.09.3 but they should be compatible with RDKit 2018.09 as well. They are not compartible with RDKit 2017.09 (due to different SMILES canonicalization). All databases contain data for contexts of radius 1, 2 and 3.
- replacements02_sa2.db.gz - database created from ChEMBL v22 structures which contain only organic atoms (C,N,O,S,P,F,Cl,Br,I) and have maximum synthetic accessibility score (SAScore) 2
- replacements02_sc2.db.gz - database created from ChEMBL v22 structures which contain only organic atoms (C,N,O,S,P,F,Cl,Br,I) and have maximum synthetic complexity score (SCScore) 2
- replacements02_sc2.5.db.gz - database created from ChEMBL v22 structures which contain only organic atoms (C,N,O,S,P,F,Cl,Br,I) and have maximum synthetic complexity score (SCScore) 2.5
Results of Guacamol tests for implemented CReM-based stochastic search and reference approaches
| task | SMILES LSTM* | SMILES GA* | Graph GA* | Graph MCTS* | CReM |
|---|---|---|---|---|---|
| Celecoxib rediscovery | 1.000 | 0.732 | 1.000 | 0.355 | 1.000 |
| Troglitazone rediscovery | 1.000 | 0.515 | 1.000 | 0.311 | 1.000 |
| Thiothixene rediscovery | 1.000 | 0.598 | 1.000 | 0.311 | 1.000 |
| Aripiprazole similarity | 1.000 | 0.834 | 1.000 | 0.380 | 1.000 |
| Albuterol similarity | 1.000 | 0.907 | 1.000 | 0.749 | 1.000 |
| Mestranol similarity | 1.000 | 0.79 | 1.000 | 0.402 | 1.000 |
| C11H24 | 0.993 | 0.829 | 0.971 | 0.410 | 0.966 |
| C9H10N2O2PF2Cl | 0.879 | 0.889 | 0.982 | 0.631 | 0.940 |
| Median molecules 1 | 0.438 | 0.334 | 0.406 | 0.225 | 0.371 |
| Median molecules 2 | 0.422 | 0.38 | 0.432 | 0.170 | 0.434 |
| Osimertinib MPO | 0.907 | 0.886 | 0.953 | 0.784 | 0.995 |
| Fexofenadine MPO | 0.959 | 0.931 | 0.998 | 0.695 | 1.000 |
| Ranolazine MPO | 0.855 | 0.881 | 0.92 | 0.616 | 0.969 |
| Perindopril MPO | 0.808 | 0.661 | 0.792 | 0.385 | 0.815 |
| Amlodipine MPO | 0.894 | 0.722 | 0.894 | 0.533 | 0.902 |
| Sitagliptin MPO | 0.545 | 0.689 | 0.891 | 0.458 | 0.763 |
| Zaleplon MPO | 0.669 | 0.413 | 0.754 | 0.488 | 0.770 |
| Valsartan SMARTS | 0.978 | 0.552 | 0.990 | 0.04 | 0.994 |
| Deco Hop | 0.996 | 0.970 | 1.000 | 0.590 | 1.000 |
| Scaffold Hop | 0.998 | 0.885 | 1.000 | 0.478 | 1.000 |
| total score | 17.341 | 14.398 | 17.983 | 9.011 | 17.919 |
Citation:
Polishchuk, P., CReM: chemically reasonable mutations framework for structure generation. Journal of Cheminformatics 2020, 12, (1), 28. - https://doi.org/10.1186/s13321-020-00431-w