
Chemoinformatics tools
Institute of Molecular and Translational Medicine, Palacky University
Simplex representation of molecular structure (SiRMS):
Simplex - tetratomic fragments of fixed composition, structure, chirality and symmetry
Simplex descriptor - number of identical simplexes in a molecule
Example of 2D simplexes generation for formic acid
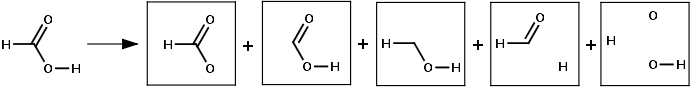
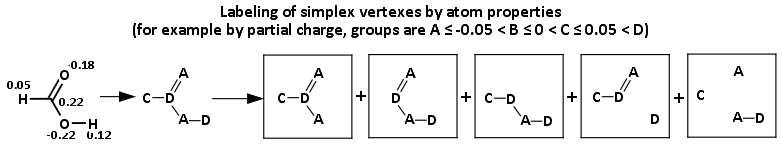
Advantages:
- vertexes in simplexes can be labeled by different atomic properties like lipophilcity, partial charge, donor/acceptor of H-bond, etc;
- simplexes describe individual compounds, multi-fragment compounds and mixtures of chemical compounds;
- simplex descriptors are interpretable by design.
We have two implementations of simplex descriptors (they are incompartible with each other)
| HiT-QSAR | SiRMS.py | |
|---|---|---|
| License | Available on request (write a letter to Prof. Kuz’min) | GPL-3 (Github page) |
| OS | Windows | Windows, Linux |
| 1D simplexes | Yes | No |
| 2D simplexes | Yes | Yes |
| 3D simplexes | Yes | No |
| Mixtures | Yes | Yes |
| Quasi-mixtures | Yes | Yes |
| Chemical reactions | No | Yes |
| Graphical interface | Yes | No |
| Atom labeling by | Element, sybyl type, lipophilicity, partial charge, refraction, donor/acceptor of H-bond, electronegativity, van der Vaals attraction/repulsion, etc | Element + user-defined |
| Multi-core support | No | Experimental |
| Speed of calculation | Slower | Faster |